Wild relatives play a vital role in the genetic improvement of crops. The process of intentionally introducing chromosomal segments from wild relatives into crops through hybridization is known as distant hybridization. Wheat is one of the major staple crops most receptive to the introgression of alien chromosomes.
The genetic and molecular mechanisms by which wild relatives improve wheat agronomic traits have long been a key research focus. While numerous genes controlling important traits in wheat have been identified as originating from wild relatives, and large-scale sequencing efforts have systematically cataloged these introgressed fragments in the wheat genome, the regulatory patterns of gene expression within these fragments remain poorly understood.
Recently, the research group of HE Fei from Laboratory of Advanced Breeding Technologies at the Institute of Genetics and Developmental Biology (IGDB), Chinese Academy of Sciences (CAS), in collaboration with the team of LING Hong-Qing (IGDB, CAS / Yazhouwan National Laboratory), proposed a novel strategy to quantify the expression abundance of introgressed fragments by utilizing public pan-genome resources to construct a reference gene set that includes sequences from Triticeae relatives, thereby maximizing the quantification of transcripts from these alien segments.
Using seedling transcriptome data from a wheat germplasm population, the research constructed an expression quantitative trait locus (eQTL) map for genes derived from Triticeae relatives. The findings reveal that, after being incorporated into the wheat genome, the expression of genes from wild relatives is generally suppressed. However, stress-responsive genes tend to exhibit more active expression.
By integrating multi-year, multi-location field trial data, the study used eQTL map to predict candidate genes for agronomic traits identified through Genome-Wide Association Studies (GWAS). These candidate genes were encoded not only by the standard 'Chinese Spring' reference genome but also by diploid and tetraploid wild relatives, providing a crucial foundation for gene cloning and functional characterization.
This work titled "Population-scale gene expression analysis reveals the contribution of expression diversity to the modern wheat improvement" was published in
Nature Communications (
Doi:10.1038/s41467-025-66100-4) on December 15, 2025.
This research was supported by the National Key Research and Development Program of China, the National Natural Science Foundation of China, and the Yazhouwan National Laboratory.
Figure. Joint eQTLs of the pan-gene atlas and GWAS, TWAS, and SMR analysis of 34 field agronomic traits and 8 Bgt isolate infection phenotypes in wheat. (Image by IGDB)
Contact:
Dr.HE Fei
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Email: fhe@genetics.ac.cn
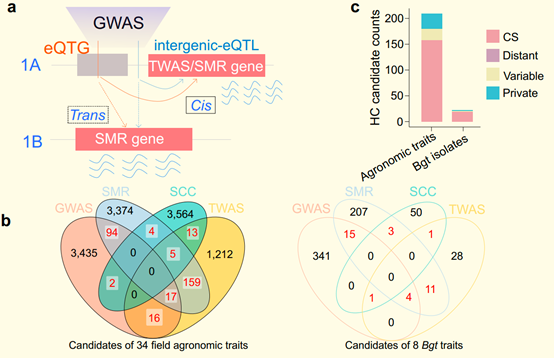 Figure. Joint eQTLs of the pan-gene atlas and GWAS, TWAS, and SMR analysis of 34 field agronomic traits and 8 Bgt isolate infection phenotypes in wheat. (Image by IGDB)Contact:Dr.HE FeiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: fhe@genetics.ac.cn
Figure. Joint eQTLs of the pan-gene atlas and GWAS, TWAS, and SMR analysis of 34 field agronomic traits and 8 Bgt isolate infection phenotypes in wheat. (Image by IGDB)Contact:Dr.HE FeiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: fhe@genetics.ac.cn CAS
CAS
 中文
中文




.png)
